Science, your chance to use all that time spent gaming for the greater good. Source Ars Technica/ Blogger ref Universal Debating Project
In the early 1950s, just as rock ‘n’ roll was hinting at social change, the first video games were quietly being designed in the form of technology demonstrations—and a scientist was behind it. In October 1958, Brookhaven National Laboratory physicist William Higinbotham created Tennis for Two. Despite graphics that are ridiculously primitive by today’s standards, it has been described as the first video game in history.
Higinbotham was inspired by the government research institution’s Donner Model 30 analog computer, which could simulate trajectories with wind resistance, and the game was designed for display at an annual public exhibition. Although his purpose in creating the game was rather academic, Tennis for Two turned out to be a hit at the three-day exhibition, with thousands of students lining up to see the game.
At first glance, today's video gamers and scientists might appear to be worlds apart. But starting with Tennis for Two, video games have quietly and consistently been within the purview of academic study. Each generation of gamers has seen new titles created at various research institutions in order to explore programming, human-computer interaction, and algorithms. Lesser-known chapters of history reveal these two worlds are not as far apart as you might think.
From Spacewar! to Foldit
After Tennis for Two's success in the late 1950, computing technology rapidly improved over the next few years. As computers became smaller and faster, more people obtained access to them. As a result, programmers began to create games for nonacademic purposes, leading up to 1962 and the “birth” of Spacewar!, the first digital computer game available outside a research institute and created solely for entertainment purposes.
During the rest of the ‘60s and early ‘70s, numerous computer games were created, and ambitious programmers saw a profitable industry rise as the audience for video games grew. Meanwhile, the separation of academia from the video game industry was becoming more and more apparent. With the release of Space Invaders in 1978 and the introduction of vector display technology a year later, a golden age of arcade video games began, one that would reach its peak with the release of Pac-Man in 1980.
In the 1990s, as advanced video game consoles entered the homes of hundreds of millions of people worldwide, the breach between the scientific/academic world and the video game industry appeared to be deeper than ever before, and possibly permanent.
Academics looked at video games and showed that they can improve a gamer’s creative thinking, teamwork skills, hand-eye coordination, problem solving, and memory (although numerous scholars have also focused on what they perceive as gaming’s negatives). But games didn’t have any apparent real-world usefulness for anyone but the gamer. That’s been changing in recent years, though, thanks to the release of online games that are not solely for entertainment purposes.
Foldit time.
A key event driving the shift came in 2008 with the release of Foldit, a revolutionary online game that enables its players to contribute to significant scientific research. Foldit was developed as part of an experimental research project conducted by the University of Washington’s Center for Game Science, in collaboration with the UW Department of Biochemistry. The Foldit website was crystal clear about the game’s purpose:
Foldit attempts to predict the structure of a protein by taking advantage of humans’ puzzle-solving intuitions and having people play competitively to fold the best proteins. Since proteins are part of so many diseases, they can also be part of the cure. Players can design brand-new proteins that could help prevent or treat important diseases.
In a virtual contest that pitted gamers against the best-known computer program designed for the task, gamers didn’t fail to impress. Only two years after Foldit’s release, more than 57,000 players were already providing useful results that matched or outperformed algorithmically computed solutions. In 2011, a team of Foldit gamers needed just 10 days to figure out the detailed molecular structure of an enzyme from an AIDS-like virus found in rhesus monkeys, a structure that had eluded experts for more than a decade.
In January 2012, Foldit gamers would accomplish the first crowdsourced redesign of a protein, an enzyme that catalyzed a reaction widely used in synthetic chemistry to synthesize everything from drugs to pesticides. Enzymes that catalyze Diels-Alder reactions have been elusive, so the achievement of Foldit gamers is significant.
An early version of that enzyme was made by a group of scientists including David Baker, a Ph.D. and protein research scientist at the University of Washington and founder of the Foldit project. They computationally designed the enzyme from scratch but soon discovered its potency wasn’t all they’d hoped for. Foldit players re-engineered the enzyme by adding 13 amino acids, increasing its activity by more than 18 times. How did they manage such an incredible feat? Thanks to Foldit’s gameplay, players could explore more radical changes to the protein than typical algorithms allow.
As Baker continued to look for useful targets to set Foldit players on, other researchers started to tap into the love some players have for games that solve complex scientific puzzles, people who would never have access to a lab under normal circumstances.
Listing image by Henrik Sorensen / Getty Images
JUMP TO ENDPAGE 1 OF 2
Phylo
Scientific gaming this decade
In 2010, while Foldit was receiving positive feedback from other researchers, scientists at Carnegie Mellon and Stanford University were developing EteRNA. Similar in concept to Foldit, EteRNA challenges its players to get RNA into a target shape. RNA plays a significant role in building proteins and modulating genes; like DNA, it’s made of four different types of nucleotide bases. To clear a level in EteRNA, the player has to switch these nucleotide bases, which changes the RNA’s configuration and changes its stability. Once a player reaches a certain level, they get the chance to play in the lab section of the game, where scientists give the best players the chance to create all-new RNA shapes. The highest ranked of all designs are then synthesized in a Stanford biochemistry lab.
Eyewire is another “game with a purpose” in which ordinary people get a chance to help science more than they could ever imagine. It was released in December 2012 and since then has brought together more than 200,000 players from over 150 countries. The mind behind this browser-based game is neuroscience expert Sebastian Seung, formerly of MIT and now at Princeton. Seung had the idea of gamifying his demanding scientific research and recruiting video game players—who most likely have nothing to do with neuroscience—to figure out how tangled sets of neurons connect to one.
If you want to excel at Eyewire you have to earn points by solving complex 3D puzzles. But the main difference from the other games is that the data you produce during gameplay won’t just put you higher on the scoreboard—it will one day be used by scientists to map the human brain (for now, players focus on mice and zebrafish to start).
If you happen to be a big fan of Candy Crush, Bejeweled, and other iconic gem-swapping puzzle games, Phylo is the right choice for you. Since the end of 2010, Phylo has been offering its players the opportunity to solve pattern-matching puzzles while playing a Tetris-like game. The goal of Phylo’s designers is to demonstrate how an NP-hard computational problem* can be embedded in a video game that draws in gamers without any significant scientific background. The twist to this game is that the data used has already been run through computer algorithms, so the gamers will be optimizing the algorithms that processed it.
Not all scientific games must focus on subatomic levels.
If virtual puzzles bore you and you’re more into adventure games, there’s one with a purpose will probably entertain you: Forgotten Island. Created by students at Syracuse University, this is another game that offers people the chance to participate in scientific research, in this case by searching for food and supplies in unwelcome forests, burning volcanoes, and barren docks.
The plot of Forgotten Island is absorbing yet simple. The player takes on the role of a lost scientist suffering from memory loss on a strange island. A catastrophic explosion has destroyed pretty much everything on the island—including a biology lab—except an enigmatic robot named DOC73R-CY3N53 who ends up being your antagonist. The robot will tell you the story of the island and then order you to reclassify his specimens and rebuild the lab.
Forgotten Island smoothly attempts to integrate taxonomic classification into the larger narrative of the game’s story. The player will interact with other characters while exploring the island and has to reclassify images from the destroyed laboratory, which are scientific photographs of living organisms collected by biologists from around the world. As a player, you will use an invention called the “Atomic Classifier” to sort the scattered biology lab photos. For each classification, you will be rewarded a small amount of game money that you can use to purchase equipment from ED-D, a childlike robot who runs the island’s only store.
It is up to the player to decide when and where to use the Atomic Classifier, but classifications are necessary to make progress in the game. Nearly 160 photos have to be classified during gameplay in order to defeat DOC73R-CY3N53.
Another fun game with similar gameplay is Happy Match. Ideal for determined observers who don’t let anything go unnoticed, Happy Match players take part in scientific research by classifying photos of animal, plant, and insect species. Scientists have designed the questions based on information they want to know. Each time someone plays, he or she is given five or 10 photos to work with. In each round, the player is asked a question about the specimens in the photos—their color, shape, and so on. By answering even simple questions, you help scientists; as you progress, the questions get progressively more complex.
Having read some reviews of this game (and tried it myself), it’s clear the gameplay isn’t really all that compelling for long. It’s not clear whether the goal is to get gamers to help out biologists, or if the developers are conducting some sort of experiment focusing on their willingness to stick with the game despite this.
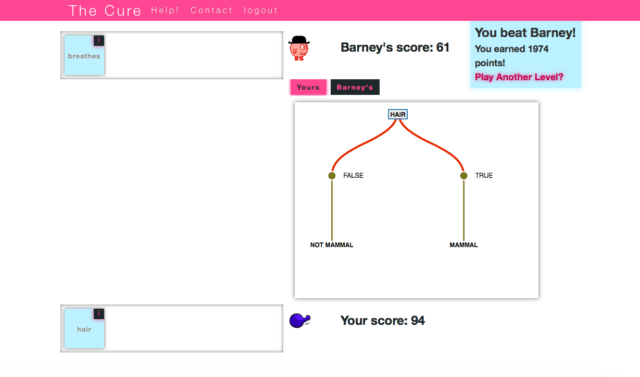
Enlarge / It's a Sunday, but we're still in love with The Cure.
Arguably one of the most ambitious computer games ever developed, The Cure attempts to discover better indicators to use in breast cancer diagnosis. Gameplay is simple: The Cure is a biology-based virtual card game in which gamers are required to assemble a “hand” of five genes from a set of 25 genes preselected for their importance to a specific type of cancer. The gene set that eventually wins is the one that produces the best predictive model of breast cancer diagnosis, as determined by a cross-validation statistical analysis.
The Cure is definitely not a game you should take lightly, as it requires a great amount of time, observation, and research, although it all serves an important purpose. One could argue that The Cure is the perfect example of how citizen science can be used to exploit the hundreds of millions of “human-brain peta-flops”** that are spent on games each day.
Whether these games mark the start of a trend depends in part on how scientists view gaming culture. While games have often been dismissed as trivial, the first video games were designed for academics. The scientific world’s recent re-approach to gamers may be a sign that academics have rediscovered the value of a good game.
Both sides, scientists and gamers, appear to be realizing how effective their collaboration can be for society. Who knows? Maybe when future historians reinvestigate the 1950s, rock ‘n’ roll might not be the main focus anymore. Instead, they may mark it as the decade when video gaming started quietly, but only a few decades later would begin to transform the world of science.
Theo is a lawyer (J.D. - M.A.) who speaks four languages and writes for a living. When he’s not working or eating chocolate, he usually plays video games, reads graphic novels, or investigates the culture of science fiction. He has written for Gizmodo, Ozy, Games Magazine, Mental Floss, and Ranker among others.
No comments:
Post a Comment